
dREG Service
The dREG model in the gateway predicts the location of enhancers and promoters using PRO-seq, GRO-seq, or ChRO-seq data. The server takes as input bigWig files provided by the user, which represent PRO-seq signal on the plus and minus strand. The gateway uses a pre-trained dREG model to identify divergent transcript start sites and impute the predicted DNase-I hypersensitivity signal across the genome. The current dREG model works in any mammalian organism.
Registered users need only upload experimental data in the required format and push the start button. Once the job is finished, the user will be notified by e-mail. Results can be downloaded to the user’s local machine, or viewed in the Genome Browser via the handy trackhub link.
Use the Danko lab's mapping pipeline (here) to prepare bigWig files from fastq files or convert BAM files of mapped reads to bigWig (here).
See our documentation, FAQ, GitHub, dREG paper, or dREG protocol for additional questions.
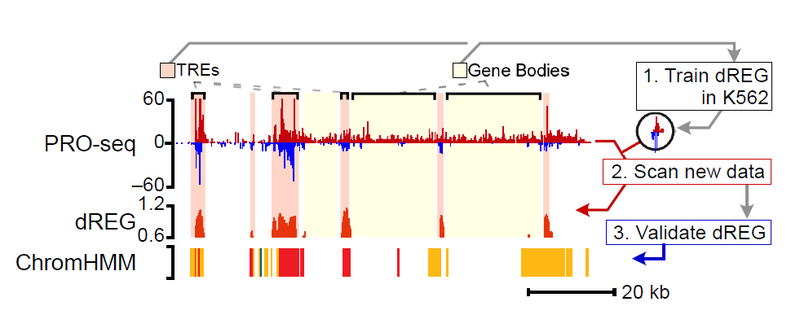
dREG Service
PGA is a science gateway built with the Airavata API. You can reference PGA as you integrate Airavata into your own gateway, or you can create your gateway on top of PGA by cloning it at the link below. PGA is known to work well in the Chrome, Firefox, and Internet Explorer browsers.

Shield
Shield from complexity
of resources

Fast
Accelerate scientific
discovery
Distributed
Access distributed
data resources
Science Gateway Research Team
The Science Gateways Research Center researches, develops, and operates science gateways in collaboration with many clients and partners. Contact us for help building science gateways and deploying advanced software cyberinfrastructure for your community.
Mission
We develop and use open source software to create user tools and environments that help scientific communities do what they could not otherwise do. Cyberinfrastructure software systems must be designed and developed strategically, not just from the requirements of scientists but from deeper understanding of core principles of cloud-scale distributed systems.
Values
The Science Gateways Research Center’s core principle is that producing world-class eScience systems requires a cooperative balance between understanding the needs and goals of scientific collaborators, undertaking core eScience research ourselves, and understanding how to operate science gateway software at scale. Open community software that follows the Apache Software Foundation principles is central to this mission.






